-
Notifications
You must be signed in to change notification settings - Fork 101
CH.1.1.1 with S:Q613H [197 sequences, United Kingdom and Sweden] #1753
New issue
Have a question about this project? Sign up for a free GitHub account to open an issue and contact its maintainers and the community.
By clicking “Sign up for GitHub”, you agree to our terms of service and privacy statement. We’ll occasionally send you account related emails.
Already on GitHub? Sign in to your account
Comments
|
128 sequences now. |
|
148 sequences now. |
|
I am having trouble finding these (currently) 148 sequences on the tree. To me it looks like DV.1 with G23401T (S:Q613H), and then the next node rootward has C29067T (N:T265I), but on usher this defines a clade of 604 DV.1 sequences, not 148 CH.1.1.1 sequences. On the Nextstrain tree linked to in this issue, there are 489 descendants off the node with S:Q613H, again far more than 148. |
|
@InfrPopGen I am confident that this branch exists, it is CH.1.1.1+Q613H (not DV.1), which can be found in the Nextstrain tree, a branch under DV.1, which had a sequence count of 148 in the last update of the cov-spectrum. |
Thank you @Outpfmance |
|
@InfrPopGen Yes, it does look that way, so should it be named CH.1.1.1.5 (DV.5) or something? Maybe there should be a discussion? |
|
cov-spectrum has just been updated, it now has 197 sequences |
|
Without having looked at it - this could well be homoplasy or artefact, needs to be looked at more closely The query given above doesn't contain a wildcard, so it queries CH.1.1.1 only, none of the DV.1 - that's not best practice as Nextclade demarkations are fuzzy - that's fine for wildcard searches, but not for exclusion purposes. |
|
There's this DV.1 sibling, on the branch with DV.3 (25721T). Small UK/Australia (60). Sequences should be CH.1.1.1 There's a CH.1.1 on C193T, C21811T branch with CH.1.1.5 etc. Mostly Sweden and Germany, 124 sequences, could be designated Then there is a 21811 branch which has quite some interesting sublineages, one is 613H and 215G. I'll designate the whole branch, the sublineages can be taken care of later if need be. That should clear it all up. @Outpfmance as you can see it's not as easy as making a query on covspectrum - there are all sorts of things that need to be checked - it helps if you include screenshots of the lineages, not just links, as the link to Usher expires. Also the Nextclade reference tree is often useful to know where on the tree you are (it's sometimes not 100% correct when there's homoplasy but can be a good guide, as in this case, see screenshot above, here's the link: https://next.nextstrain.org/staging/nextclade/sars-cov-2/21L?branchLabel=aa) |



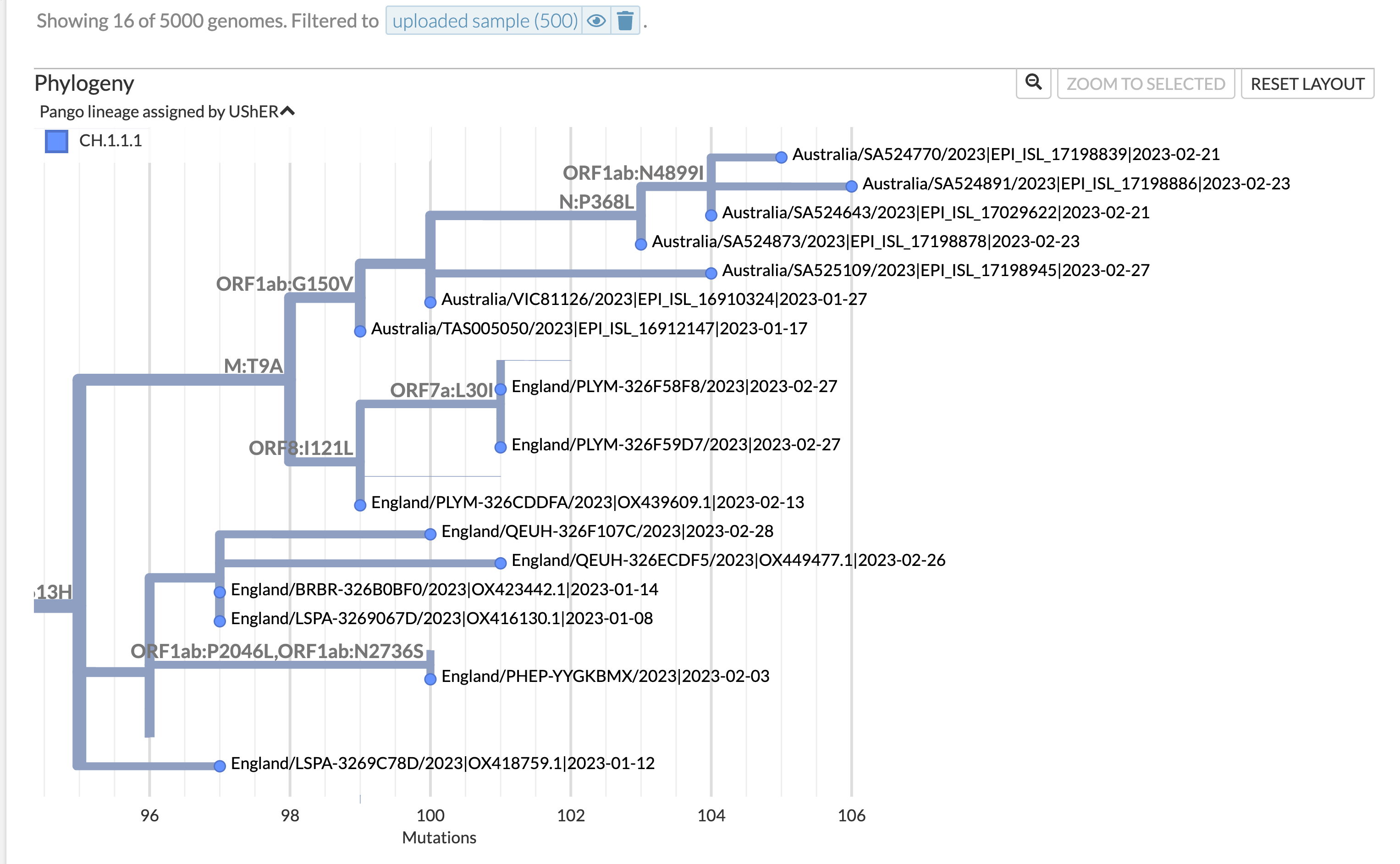

This is another branch with the Q613H mutation, which is already present in a large proportion in DV.1 (CH.1.1.1.1), as well as in the ancestor of DV.1, CH.1.1.1, which is not as large as in DV.1, but is of concern.
And in recent times, the relative growth advantage of this variant has increased.
Cov-spectrum:
https://cov-spectrum.org/explore/World/AllSamples/from%3D2023-01-01%26to%3D2023-03-01/variants?aaMutations=s%3A613h&nextcladePangoLineage=CH.1.1.1&
Nextstrain: https://nextstrain.org/fetch/genome.ucsc.edu/trash/ct/singleSubtreeAuspice_genome_30e5b_85b1b0.json?f_userOrOld=uploaded%20sample
The text was updated successfully, but these errors were encountered: